Below is a collection of the Whelan Lab research projects. The projects are sorted by the word clouds which highlight topics of interest.
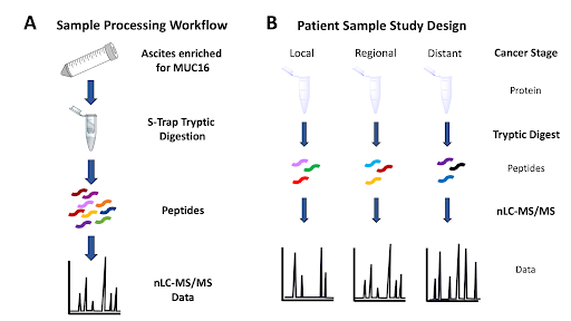
Mass spectrometry characterization of the ovarian cancer biomarker CA125 (MUC16) from patient-derived ascites
Ascites is fluid that accumulates in the peritoneal cavity of patients with advanced stage ovarian cancer. Our lab, in collaboration with Dr. Manish Patankar at the University of Wisconsin School of Public Health and Medicine, has developed a process to enrich ascites for MUC16 and characterize the product using bottom-up proteomics. MUC16 is the glycoprotein that contains the CA125 epitope; CA125 is an FDA approved biomarker for patients with ovarian cancer. We have already optimized a bottom-up proteomics workflow that uses deoxycholic acid and STraps to digest small, microgram amounts of proteins [Ref. Schuster-Little 2020]. Following digestion, proteins (including MUC16) are characterized using nano liquid chromatography tandem mass spectrometry (nLC-MS/MS). We hypothesize that MUC16 expression varies between individuals and during different stages of ovarian cancer, and that these differences will be identifiable using mass spectrometry. Additionally, we are investigating the correlation between the clinical CA125 assay and mass spectrometry detectable peptides. The CA125 epitope constitutes 1% of the total protein MUC16. We hypothesize that a mass spectrometry-based assay will detect analytically silent regions of the protein that are currently undetected using the clinical assay.
Funding: Chemistry Biochemistry Biology Interface (CBBI) Fellowship awarded to Naviya
Ascites is fluid that accumulates in the peritoneal cavity of patients with advanced stage ovarian cancer. Our lab, in collaboration with Dr. Manish Patankar at the University of Wisconsin School of Public Health and Medicine, has developed a process to enrich ascites for MUC16 and characterize the product using bottom-up proteomics. MUC16 is the glycoprotein that contains the CA125 epitope; CA125 is an FDA approved biomarker for patients with ovarian cancer. We have already optimized a bottom-up proteomics workflow that uses deoxycholic acid and STraps to digest small, microgram amounts of proteins [Ref. Schuster-Little 2020]. Following digestion, proteins (including MUC16) are characterized using nano liquid chromatography tandem mass spectrometry (nLC-MS/MS). We hypothesize that MUC16 expression varies between individuals and during different stages of ovarian cancer, and that these differences will be identifiable using mass spectrometry. Additionally, we are investigating the correlation between the clinical CA125 assay and mass spectrometry detectable peptides. The CA125 epitope constitutes 1% of the total protein MUC16. We hypothesize that a mass spectrometry-based assay will detect analytically silent regions of the protein that are currently undetected using the clinical assay.
Funding: Chemistry Biochemistry Biology Interface (CBBI) Fellowship awarded to Naviya
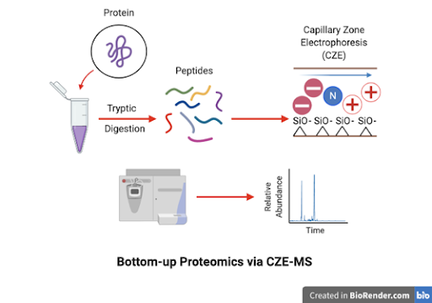
Utilizing a CZE-mass spectrometry based assay for the analysis of CA125
The detection of the glycoprotein CA125 is used in clinical settings for the diagnosis of ovarian cancer. Unfortunately, clinical assays lack specificity and sensitivity to provide accurate results because other noncancerous conditions can cause elevated CA125 levels. We hypothesize that the structures and epitopes of CA125 are detectable through capillary zone electrophoresis-mass spectrometry (CZE-MS) and liquid chromatography-mass spectrometry (LC-MS). CZE is complementary to LC, but is often underutilized in the proteomics field when compared to LC.
CZE often favors unique hydrophilic and low mass m/z peptides not detected by LC. Combined with a bottom-up proteomics approach, we use CZE-MS and LC-MS as an alternative detection method with the possibility to monitor CA125 levels prior, during, and post treatment.
The detection of the glycoprotein CA125 is used in clinical settings for the diagnosis of ovarian cancer. Unfortunately, clinical assays lack specificity and sensitivity to provide accurate results because other noncancerous conditions can cause elevated CA125 levels. We hypothesize that the structures and epitopes of CA125 are detectable through capillary zone electrophoresis-mass spectrometry (CZE-MS) and liquid chromatography-mass spectrometry (LC-MS). CZE is complementary to LC, but is often underutilized in the proteomics field when compared to LC.
CZE often favors unique hydrophilic and low mass m/z peptides not detected by LC. Combined with a bottom-up proteomics approach, we use CZE-MS and LC-MS as an alternative detection method with the possibility to monitor CA125 levels prior, during, and post treatment.
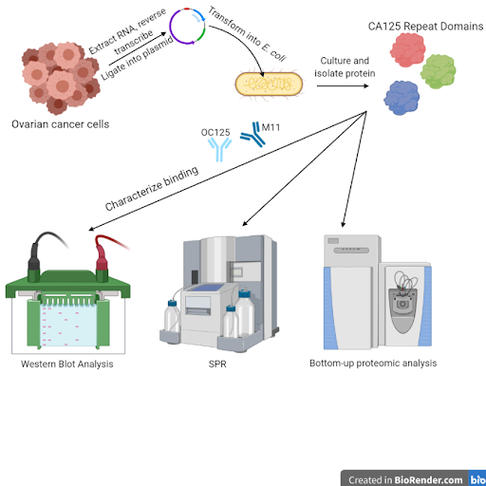
Assessment of binding affinity and epitopes of CA125 antibodies to repeat domains
Ovarian cancer progression and recurrence are clinically monitored over time with an antibody-based test for the biomarker protein CA125. However, the ways in which OC125 and M11, the two antibodies used in the test, interact with CA125 has not been fully characterized. Further understanding of the interactions between these antibodies and their target has the potential to help with assay optimization, or to develop new tests that could be used for screenings for earlier diagnosis. This project aims to express repeat domains of the CA125 protein individually in E. coli, and to assess antibody binding affinity to different repeats using a variety of techniques, including Western blot analysis, surface plasmon resonance (SPR), liquid chromatography-tandem mass spectrometry (LC-MS/MS), and capillary zone electrophoresis-tandem mass spectrometry (CZE-MS/MS). We will also use bottom-up proteomic analysis on strong binding repeats cross-linked with antibodies in order to map the CA125 epitope(s), or sites of antibody binding, of both OC125 and M11.
Funding: T.E.A.L. Medical Research and Awareness Program
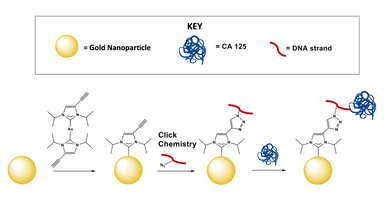
Using SERS and N-heterocyclic carbenes to detect CA125
In collaboration with Jon Camden (University of Notre Dame) and David Jenkins (University of Tennessee) we aim to use Surface Enhanced Raman Spectroscopy (SERS) to detect and quantify CA125. A SERS assay would enable label-free detection of CA125. Traditionally, thiols are used to link a substrate to a gold nanoparticle. We aim to use NHCs as an alternative to thiols. We will use a CA125 aptamer, selected in-house, to couple to the gold nanoparticle surface. Following attachment, we will test our assay in biologically relevant media.
Funding: Institute for Precision Health at the University of Notre Dame
In collaboration with Jon Camden (University of Notre Dame) and David Jenkins (University of Tennessee) we aim to use Surface Enhanced Raman Spectroscopy (SERS) to detect and quantify CA125. A SERS assay would enable label-free detection of CA125. Traditionally, thiols are used to link a substrate to a gold nanoparticle. We aim to use NHCs as an alternative to thiols. We will use a CA125 aptamer, selected in-house, to couple to the gold nanoparticle surface. Following attachment, we will test our assay in biologically relevant media.
Funding: Institute for Precision Health at the University of Notre Dame
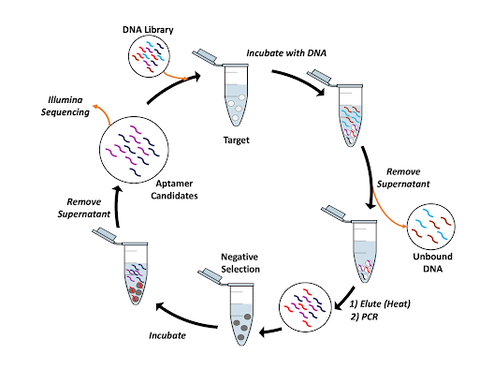
Selection of aptamers for the peptide hormone oxytocin
Oxytocin is a naturally occurring peptide hormone that plays a role in social bonding, sexual reproduction, childbirth, and postpartum recovery. A synthetic version of oxytocin is commonly administered to mothers during and after childbirth. The drug is temperature sensitive and degrades quickly when exposed to high temperatures; degraded oxytocin becomes inactive and is ineffective due to loss of chemical activity. When a dose of inactive oxytocin is administered to mothers after childbirth, there is an increased risk for hemorrhaging and maternal death. We aim to develop an affordable, easy to use test using paper analytical devices (PADs) to determine if commercially available oxytocin has degraded. Our ultimate goal is to distribute these PADs to resource limited environments where traditional quality control testing is cost-prohibitive. The Whelan lab is selecting aptamers for oxytocin and its common degradation products using a magnetic-bead based approach. Following aptamer selection and characterization, our lab will work in collaboration with the Lieberman lab to develop a PAD suitable for use in resource limited environments. We hypothesize that this test will be simple and fast for medical staff to use to determine if a dose or vial of oxytocin is biologically active and safe to administer to women.
Funding: The Indiana Clinical and Translational Sciences Institute (CTSI) through Disease Diagnostic INventors Challenge (DDIN)
Oxytocin is a naturally occurring peptide hormone that plays a role in social bonding, sexual reproduction, childbirth, and postpartum recovery. A synthetic version of oxytocin is commonly administered to mothers during and after childbirth. The drug is temperature sensitive and degrades quickly when exposed to high temperatures; degraded oxytocin becomes inactive and is ineffective due to loss of chemical activity. When a dose of inactive oxytocin is administered to mothers after childbirth, there is an increased risk for hemorrhaging and maternal death. We aim to develop an affordable, easy to use test using paper analytical devices (PADs) to determine if commercially available oxytocin has degraded. Our ultimate goal is to distribute these PADs to resource limited environments where traditional quality control testing is cost-prohibitive. The Whelan lab is selecting aptamers for oxytocin and its common degradation products using a magnetic-bead based approach. Following aptamer selection and characterization, our lab will work in collaboration with the Lieberman lab to develop a PAD suitable for use in resource limited environments. We hypothesize that this test will be simple and fast for medical staff to use to determine if a dose or vial of oxytocin is biologically active and safe to administer to women.
Funding: The Indiana Clinical and Translational Sciences Institute (CTSI) through Disease Diagnostic INventors Challenge (DDIN)
Thermal oscillation SELEX for the ovarian cancer biomarker HE4
We aim to select aptamers for the ovarian cancer biomarker HE4 using a novel Systematic Evolution of Ligands by EXponential Enrichment (SELEX) selection method, thermal oscillation SELEX. Aptamers are single stranded DNA or RNA that are traditionally selected using SELEX. During the SELEX process a pool of 10^15-10^16 random DNA sequences are incubated with a target and narrowed down to a select pool of high affinity aptamer candidates. We have developed a new magnetic bead-based SELEX that uses thermal oscillation to disrupt initial weak-binding aptamer-protein interactions. Upon multiple cycles of thermal oscillation, weak-binding aptamers will dissociate, enabling potentially stronger aptamer-protein complexes to be made. Through this process, we have selected a set of high affinity aptamers (determined by bulk affinity) with only 2 rounds of selection. Further steps will be to determine the affinity of individual aptamers using fluorescence anisotropy (FA), affinity probe capillary electrophoresis (APCE), and surface plasmon resonance (SPR).
We aim to select aptamers for the ovarian cancer biomarker HE4 using a novel Systematic Evolution of Ligands by EXponential Enrichment (SELEX) selection method, thermal oscillation SELEX. Aptamers are single stranded DNA or RNA that are traditionally selected using SELEX. During the SELEX process a pool of 10^15-10^16 random DNA sequences are incubated with a target and narrowed down to a select pool of high affinity aptamer candidates. We have developed a new magnetic bead-based SELEX that uses thermal oscillation to disrupt initial weak-binding aptamer-protein interactions. Upon multiple cycles of thermal oscillation, weak-binding aptamers will dissociate, enabling potentially stronger aptamer-protein complexes to be made. Through this process, we have selected a set of high affinity aptamers (determined by bulk affinity) with only 2 rounds of selection. Further steps will be to determine the affinity of individual aptamers using fluorescence anisotropy (FA), affinity probe capillary electrophoresis (APCE), and surface plasmon resonance (SPR).
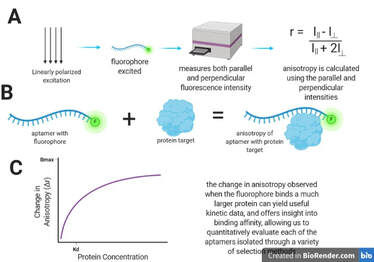
Aptamer characterization: instrumental quantitation of binding affinity
The five-year survival rate for ovarian cancer jumps dramatically if diagnosed at a local stage as opposed to after metastasis, but only a fraction of cases are diagnosed early. There are two FDA approved protein biomarkers for ovarian cancer, CA125 and HE4, but because of high false positive and false negative rates with the traditional antibody tests used to detect them, they are used to monitor the progression of the disease rather than in a diagnostic capacity. Because aptamers have several benefits over antibodies, including efficient development and batch-to-batch consistency, we argue that aptamers are a promising alternative method to develop improved diagnostic tests. Aptamers are selected for using an array of methods, both in-house in the Whelan lab and by our collaborators in the Spirito lab.
Binding affinity characterization furthers the goals of developing diagnostic assays by offering concrete comparisons to other aptamer and antibody candidates. Quantitation of binding affinity also helps to determine precisely how useful any aptamer may be to identify targets in patient samples.
A variety of techniques are used. Fluorescence anisotropy interrogates a sample with linearly polarized light, and then uses the measurement of depolarization to calculate anisotropy, which can yield information about binding kinetics and affinity. Affinity probe capillary electrophoresis separates analytes based on size and charge, and the addition of an affinity probe can be used to detect one of the analytes injected at various concentrations, offering information about binding between aptamer and protein target to form a complex. Surface Plasmon Resonance (performed on the Nicoya OpenSPR instrument) immobilizes one of the analytes on a sensor surface coated with nano-structured gold and flows the other analyte past the binding partner. This changes the refractive index, and absorbance measurements are used to measure binding activity.
Current projects include the characterization of:
-a set of DNA aptamers with CA125, selected for by the Whelan lab using One-Pot SELEX and the Galaxy bioinformatics pipeline.
-a group of RNA aptamers selected against HE4 protein by our collaborators in the Spirito lab
-a batch of DNA aptamers isolated through the use of Thermal Oscillation SELEX with HE4
The five-year survival rate for ovarian cancer jumps dramatically if diagnosed at a local stage as opposed to after metastasis, but only a fraction of cases are diagnosed early. There are two FDA approved protein biomarkers for ovarian cancer, CA125 and HE4, but because of high false positive and false negative rates with the traditional antibody tests used to detect them, they are used to monitor the progression of the disease rather than in a diagnostic capacity. Because aptamers have several benefits over antibodies, including efficient development and batch-to-batch consistency, we argue that aptamers are a promising alternative method to develop improved diagnostic tests. Aptamers are selected for using an array of methods, both in-house in the Whelan lab and by our collaborators in the Spirito lab.
Binding affinity characterization furthers the goals of developing diagnostic assays by offering concrete comparisons to other aptamer and antibody candidates. Quantitation of binding affinity also helps to determine precisely how useful any aptamer may be to identify targets in patient samples.
A variety of techniques are used. Fluorescence anisotropy interrogates a sample with linearly polarized light, and then uses the measurement of depolarization to calculate anisotropy, which can yield information about binding kinetics and affinity. Affinity probe capillary electrophoresis separates analytes based on size and charge, and the addition of an affinity probe can be used to detect one of the analytes injected at various concentrations, offering information about binding between aptamer and protein target to form a complex. Surface Plasmon Resonance (performed on the Nicoya OpenSPR instrument) immobilizes one of the analytes on a sensor surface coated with nano-structured gold and flows the other analyte past the binding partner. This changes the refractive index, and absorbance measurements are used to measure binding activity.
Current projects include the characterization of:
-a set of DNA aptamers with CA125, selected for by the Whelan lab using One-Pot SELEX and the Galaxy bioinformatics pipeline.
-a group of RNA aptamers selected against HE4 protein by our collaborators in the Spirito lab
-a batch of DNA aptamers isolated through the use of Thermal Oscillation SELEX with HE4
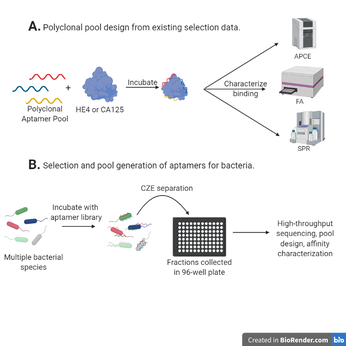
Development of polyclonal aptamer pools for both protein and bacterial targets, using both existing and newly generated selection data
Aptamers are incredibly useful alternatives to antibodies for specific detection of a biological analyte because of their low cost, uniformity, and ease of synthesis, among other properties. The potential utility of polyclonal aptamers, or multiple aptamer sequences with varying epitopes used to target one analyte, has yet to be deeply explored. We believe that polyclonal aptamers can be used to create affinity probes for complex targets, such as whole cells or variably modified proteins, and show evidence of higher bulk affinity while maintaining high pool uniformity, which is characteristic of aptamers. In order to assess this potential, polyclonal pools will be generated first from existing selection data for ovarian cancer biomarker proteins to compare to monoclonal results and optimize pool design, with binding affinity being determined using affinity probe capillary electrophoresis (APCE), fluorescence anisotropy (FA), and surface plasmon resonance (SPR). Then, aptamer selection will be performed on multiple bacterial species simultaneously using a capillary electrophoresis-fraction collector to separate these species, high-throughput sequencing will be carried out on the collected aptamers, polyclonal pools will be developed from this selection data, and binding affinity will be assessed.
Funding: Center for Bioanalytical Metrology, an NSF I/UCRC, in partnership with the Indiana Biosciences Research Institute, and the University of Notre Dame Institute for Precision Health
Aptamers are incredibly useful alternatives to antibodies for specific detection of a biological analyte because of their low cost, uniformity, and ease of synthesis, among other properties. The potential utility of polyclonal aptamers, or multiple aptamer sequences with varying epitopes used to target one analyte, has yet to be deeply explored. We believe that polyclonal aptamers can be used to create affinity probes for complex targets, such as whole cells or variably modified proteins, and show evidence of higher bulk affinity while maintaining high pool uniformity, which is characteristic of aptamers. In order to assess this potential, polyclonal pools will be generated first from existing selection data for ovarian cancer biomarker proteins to compare to monoclonal results and optimize pool design, with binding affinity being determined using affinity probe capillary electrophoresis (APCE), fluorescence anisotropy (FA), and surface plasmon resonance (SPR). Then, aptamer selection will be performed on multiple bacterial species simultaneously using a capillary electrophoresis-fraction collector to separate these species, high-throughput sequencing will be carried out on the collected aptamers, polyclonal pools will be developed from this selection data, and binding affinity will be assessed.
Funding: Center for Bioanalytical Metrology, an NSF I/UCRC, in partnership with the Indiana Biosciences Research Institute, and the University of Notre Dame Institute for Precision Health
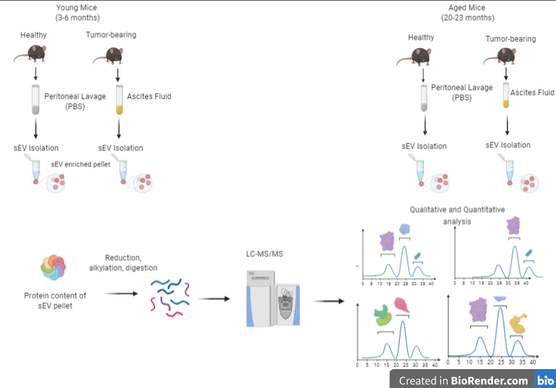
Proteomic and lipidomic analysis of small extracellular vesicles from young and aged ovarian cancer murine models
Aging is the single greatest risk factor for ovarian cancer. Older women are diagnosed more frequently and have poorer prognosis due to metastasis occurring much more rapidly than in younger women. Metastasis relies on cell-cell communication to progress. Small extracellular vesicles (sEVs) are key players in this cell-cell communication. This project aims to explore any potential differences in both the proteome and the lipidome of these sEVs via liquid chromatography coupled to tandem mass spectrometry. We hypothesize that differences in the proteomic and lipidomic profiles of young and aged sEVs help promote metastatic success in older subjects. Biofluids from young and aged murine models will undergo an in-depth characterization of both the proteome and lipidome of sEVs. Statistical and pathway analysis will be performed to explore how differences between these groups of sEVs could cause the observed increase in metastatic ability of OvCa in older women.
Aging is the single greatest risk factor for ovarian cancer. Older women are diagnosed more frequently and have poorer prognosis due to metastasis occurring much more rapidly than in younger women. Metastasis relies on cell-cell communication to progress. Small extracellular vesicles (sEVs) are key players in this cell-cell communication. This project aims to explore any potential differences in both the proteome and the lipidome of these sEVs via liquid chromatography coupled to tandem mass spectrometry. We hypothesize that differences in the proteomic and lipidomic profiles of young and aged sEVs help promote metastatic success in older subjects. Biofluids from young and aged murine models will undergo an in-depth characterization of both the proteome and lipidome of sEVs. Statistical and pathway analysis will be performed to explore how differences between these groups of sEVs could cause the observed increase in metastatic ability of OvCa in older women.